
-What Makes a Mammal a Mammal?: An Approach to Mammalian-specific Traits from the Perspective of Genomic Imprinting and Genes exapted from LTR Retrotransposons/Retroviruses-

-What Makes a Mammal a Mammal?: An Approach to Mammalian-specific Traits from the Perspective of Genomic Imprinting and Genes exapted from LTR Retrotransposons/Retroviruses-
We discovered the PEG10 gene as one of the paternally expressed imprinted genes in the course of research on genomic imprinting that is known to take place via a mammalian-specific epigenetic mechanism. PEG10 has some unique features, e.g. it: 1) resembles an LTR retrotransposon or retroviral DNA, 2) is present only in viviparous mammals, such as eutherians and marsupials, 3) plays an essential role as an endogenous gene required for placenta formation. Thus, it seems clear that PEG10 emerged by exaptation of an exogenous DNA acquired in a common ancestor of therian mammals. Moreover, we have accumulated evidence that approximately 10 such exapted SIRH/RTL genes play important roles in various aspects of eutherian development, growth and behavior.
Today, it is quite “natural” that a mammalian mother delivers one or more babies. However, it is highly likely that chance events, such as mutation and gene exaptation from exogenous DNA, underlay the emergence of mammals in the course of evolution, events that would result in viviparous reproduction as well as the highly sophisticated central nervous system (CNS). This was achieved by supplying essential placental as well as CNS genes in a mammalian-specific manner. It may be that only a very small number of external DNAs inserted into the ancestor mammalian genome came to be selected as beneficial genes by an initial series of chance events that now determine the way the current mammalian developmental system works by becoming fundamental parts of the mammalian genome.
We believe this research provides an excellent example of how genes exapted from LTR retrotransposons/retroviruses contributed to the establishment of mammals. In other words, how mammalian evolution was driven by exapted genes follows a story-line of “chance event to necessity.”
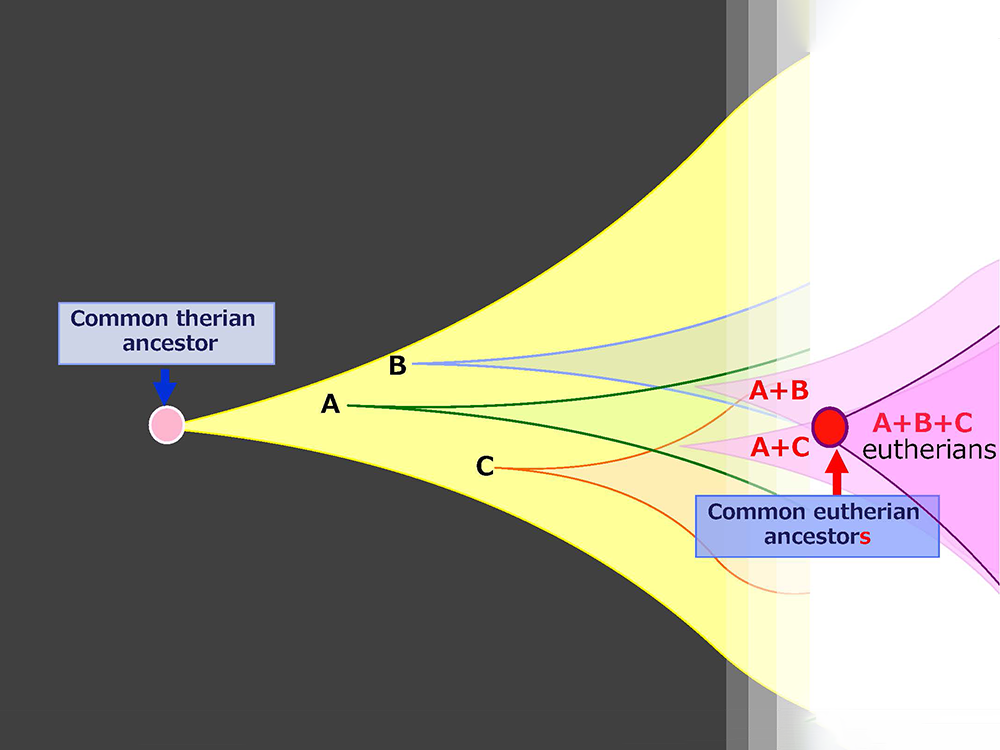
Genomic imprinting is a gene expression mechanism unique to mammals. From the perspective of genomic imprinting, we explored how viviparity—a defining reproductive trait of mammals —was established and discovered the PEG10 gene, which is essential for placentation. In the 1990s, when we embarked on a comprehensive search for imprinted genes involved in genomic imprinting, we hypothesized that some imprinted genes were newly acquired in mammals and may have contributed to the emergence of mammal-specific traits. We also speculated that the source of these new genes could be fragments of viral genes integrated into the host genome (Figure 1).
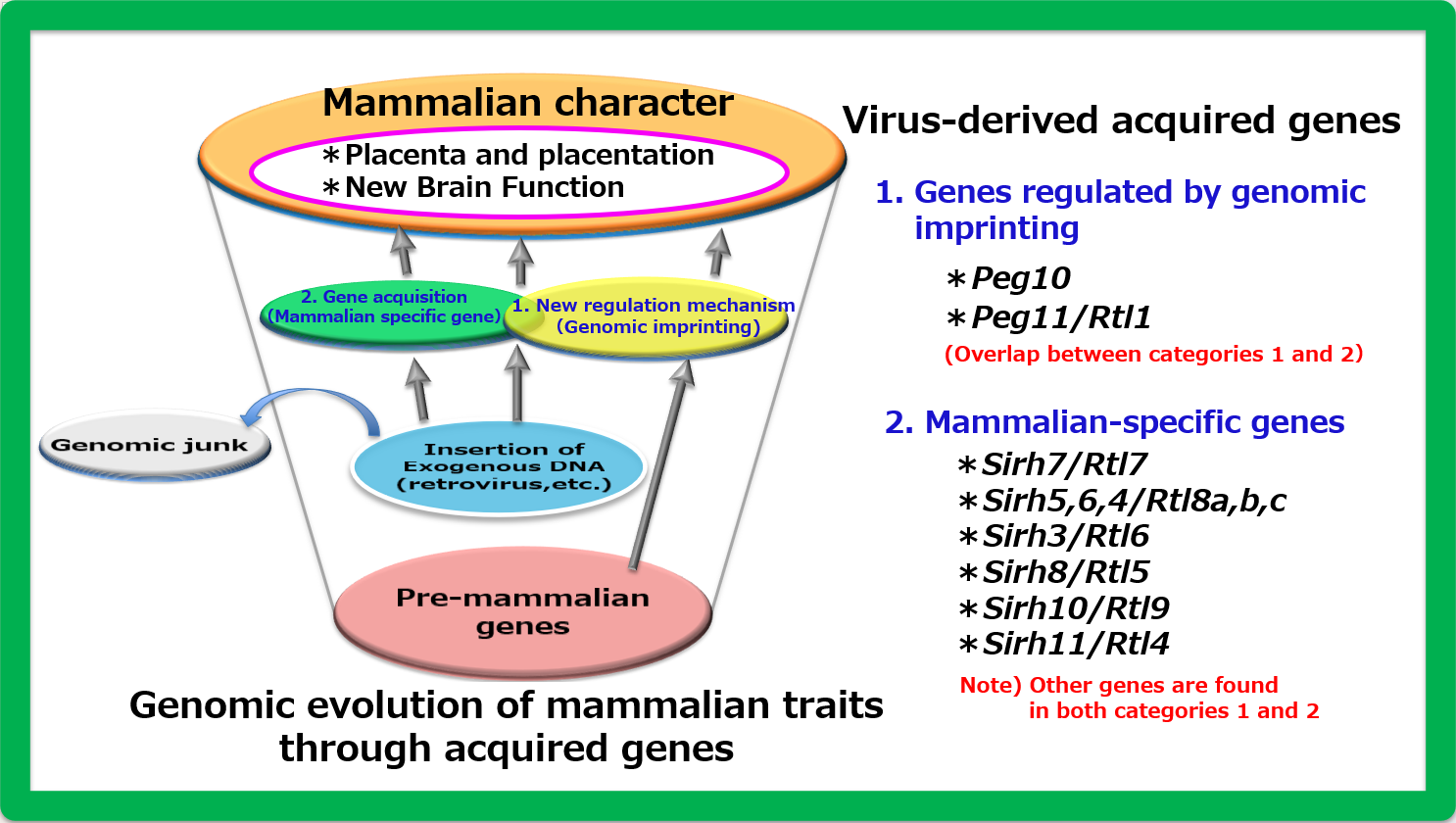
The Peg10 gene is an acquired gene derived from foreign DNA, with a sequence similar to that of a virus, and is found exclusively in viviparous mammals. Today, it seems natural and inevitable for mammals that mothers give birth to and nurture their babies. However, it turns out that the 'accidental' acquisition of the PEG10 gene through viral infections played a major role in establishing the reproductive mechanism of viviparity.
A more comprehensive search into the origins of viviparity in the mammalian genome has revealed several additional mammal-specific acquired genes, although most of them are unrelated to genomic imprinting. Nevertheless, while some of these genes —alongside PEG10—contribute to placentation, the majority are significantly associated with brain function. Since a highly advanced brain is one of the defining traits of mammals, it can be concluded that these genes have played a crucial role in shaping mammalian characteristics.
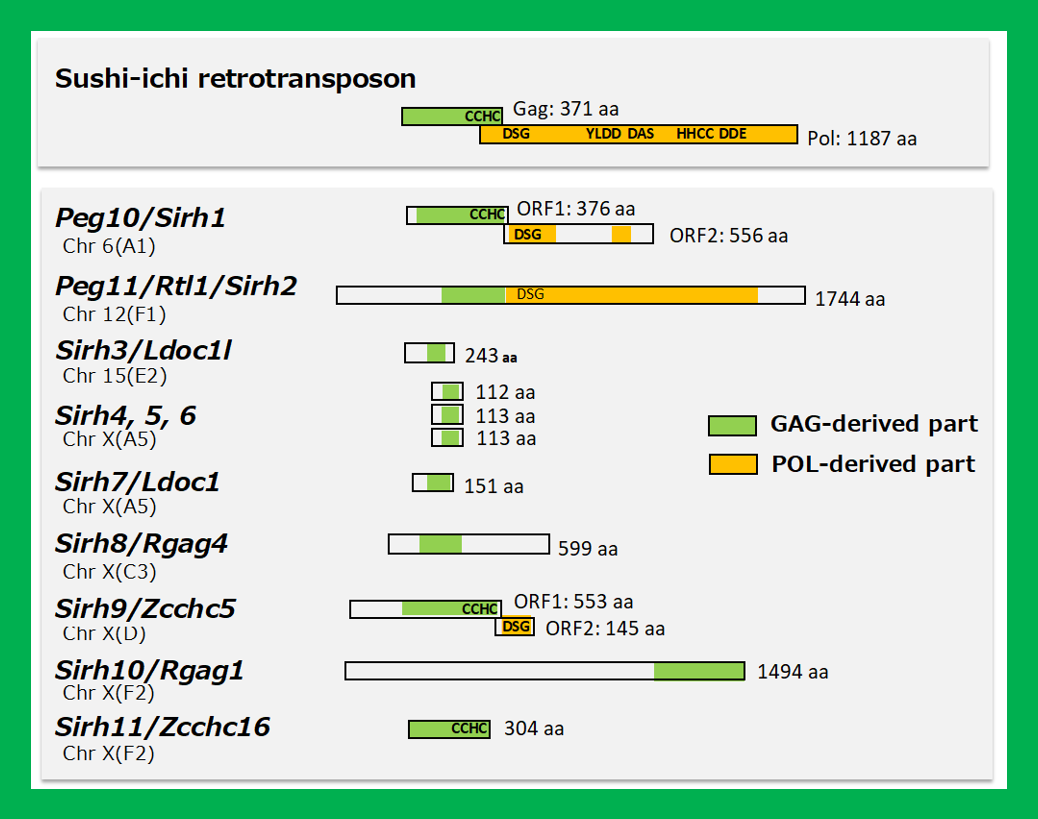
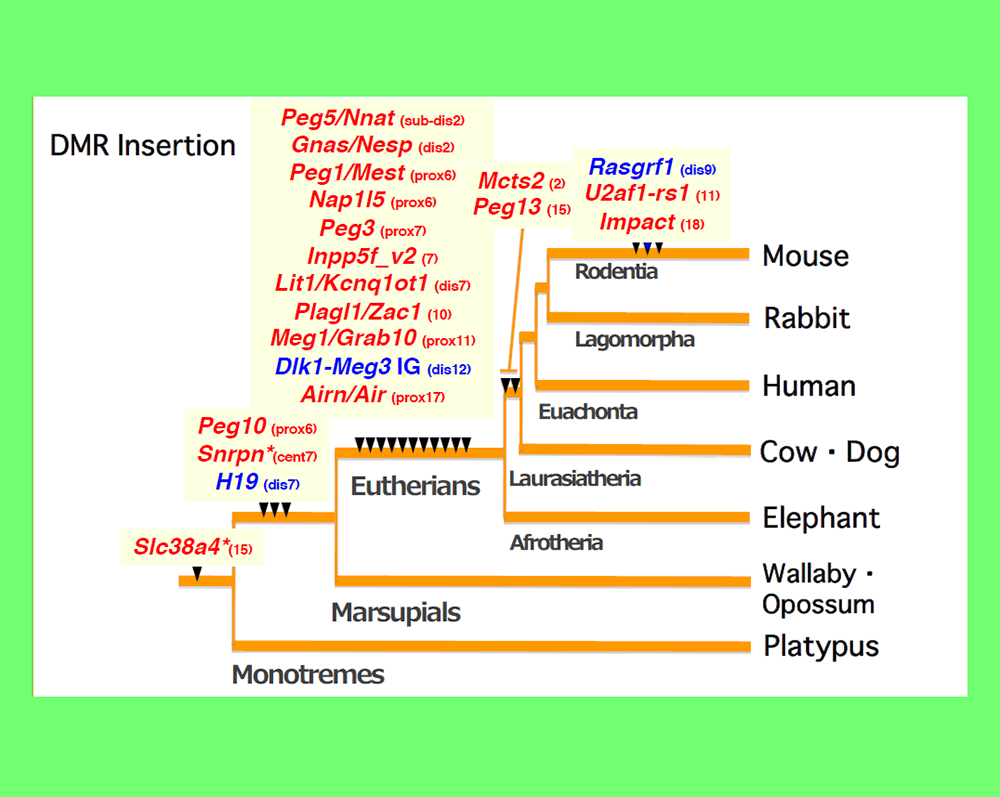
PEG10 is an acquired gene derived from the retrovirus (LTR retrotransposon) GAG and POL (Fig. 2). It is present only in therian mammals, such as eutherians, including humans and mice, as well as in marsupials such as kangaroos and koalas (Ono et al. Genomics 2001, Suzuki et al. PLoS Genet 2007), and plays an essential role in placenta formation (Ono et al. Nat Genet 2006) (Theme 4).
Another gene with an important function in the placenta is PEG11/RTL1, which was also identified in genomic imprinting studies (Charlier et al. Genome Res 2001). This acquired gene, derived from GAG and POL, is found exclusively in eutherians (Fig. 2). PEG11/RTL1 plays a crucial role in maintaining fetal capillaries in the eutherian placenta, which are essential for supplying nutrition and oxygen from the mother (Sekita et al. Nat Genet 2008, Kagami and Sekita et al. Nat Genet 2008), The acquisition of this may have enabled eutherians to have long gestation periods (Theme 5).
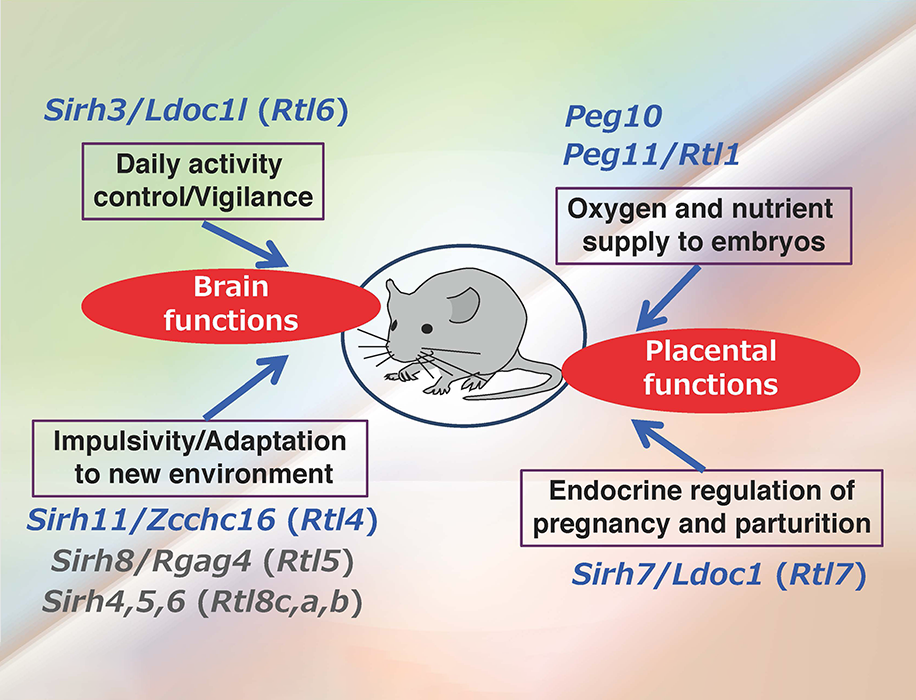
In addition to PEG10 and PEG11/RTL1, eutherians, including humans and mice, possess nine other mammalian-specific acquired genes in their lineage. These genes, including PEG10 and PEG11/RTL1, are collectively referred to as SIRH/RTL genes, as they are all derived from the same retroviral (LTR retrotransposon) GAGs (Fig. 2).
Among SIRH/RTL genes, Sirh7/Ldoc1/Rtl7 functions in the placenta; Sirh7 plays a critical role in determining the timing of delivery by regulating placental endocrine function, which is essential for the survival of pups (Naruse et al. Development 2014) (Theme 5, Spin-off 1). Furthermore, PEG10 has been found to play a vital role throughout gestation, contributing to both early placentation and the maintenance of fetal capillaries (Shiura et al. Development 2021) (Theme 10).
Regarding brain function, the SIRH/RTL genes can be classified into two groups based on their roles in neurons and microglia.
The GAG-derived genes Sirh4, Sirh5, and Sirh6 (Rtl8c, Rtl8a and Rtl8b) function in neurons (Fujioka et al. Open Biol 2025).
These three homologous genes form a cluster on the X chromosome. Mice with deletions of two of these genes (Sirh5, 6/Rtl8a, b DKO) exhibit late-onset obesity and abnormal behaviors, such as reduced activity, decreased sociability, increased apathy, and impaired maternal nursing behavior. The phenotypic similarities observed in DKO mice suggest that these genes may be linked to Prader-Willi syndrome, a genomic imprinting disorder (Theme 8).
Peg11/Rtl1, which was previously mentioned in "Creates the placenta" plays a crucial role in both placental development and brain function. In mice, deletion or overexpression of Peg11/Rtl1 results in placental abnormalities, reduced motor function and increased anxiety-like behavior. Additionally, overexpression significantly impairs spatial memory. In humans, deletion of PEG11/RTL1 is associated with Temple syndrome, while overexpression is linked to Kagami-Ogata syndrome, both of which are genomic imprinting disorders (Theme 7).
Peg10 has also been shown to play a crucial role in brain function, as demonstrated by brain-specific conditional KO mice (deletion of Peg10 exclusively in the brain) (Shiura, unpublished data). Furthermore, elevated PEG10 protein levels have been observed in iPS-differentiated neurons derived from patients with Angelman syndrome—another genomic imprinting disorder—as well as in those with amyotrophic lateral sclerosis (ALS), suggesting a potential role for PEG10 in these conditions (Pandya et al. Cell Rep Med 2021; Whitelay et al. J Biol Chem 2021; Black et al. eLife 2023).
Four genes—Sirh3/Rtl6, Sirh8/Rtl5, Sirh10/Rtl9 and Sirh11/Rtl4/Zcchc16—function in microglia, the immune cells of the brain. Among them, three genes (excluding Sirh11) are involved in protecting the brain against pathogen infections (Irie et al. Development 2022; Ishino et al. Int J Mol Sci 2023).
Innate immunity is the body's first line of defense against infections caused by bacteria, viruses and fungi. This mechanism is evolutionarily conserved across the animal kingdom. Various Toll-like receptors (TLRs) recognize pathogen-associated molecular patterns (PAMPs) and trigger an inflammatory response to counteract them. Unlike other organs, the brain relies solely on microglia for innate immunity. SIRH3 and SIRH8, proteins secreted by microglia and widely distributed throughout the brain, rapidly form complexes upon detecting their recognized PAMPs. This prevents pathogen spread and facilitates swift clearance. In contrast, SIRH10 localizes to microglial lysosomes, where it plays a role in degrading engulfed zymosan, a component of fungi cell walls. The discovery that acquired genes contribute to PAMP clearance suggests that eutherians have experienced a unique evolution of innate immune mechanisms in the brain (Theme 9).
Sirh11/Rtl4/Zcchc16, on the other hand, serves a different function. We previously reported that Sirh11/Zcchc16 KO mice exhibit impaired adaptation to novel environments, increased impulsivity and impaired short-term spatial memory (Irie et al. PLoS Genet 2015). Mutations in this gene have also been implicated in the development of autism spectrum disorders (ASD) in humans (Lim et al. Neuron 2013). In light of this, we analyzed the dynamics of the SIRH11 protein produced by this gene in the brain.
The results showed that the SIRH11 protein is secreted by microglia, remains stable during wakefulness, and responds to the neurotransmitter noradrenaline, which is released into the brain during stress (Ishino et al. Int J Mol Sci 2024) (Theme 9). This represents another evolutionarily significant adaptation in eutherians, enabling the safer and more efficient functioning of the ancient and crucial hormone, noradrenaline, in the brain."
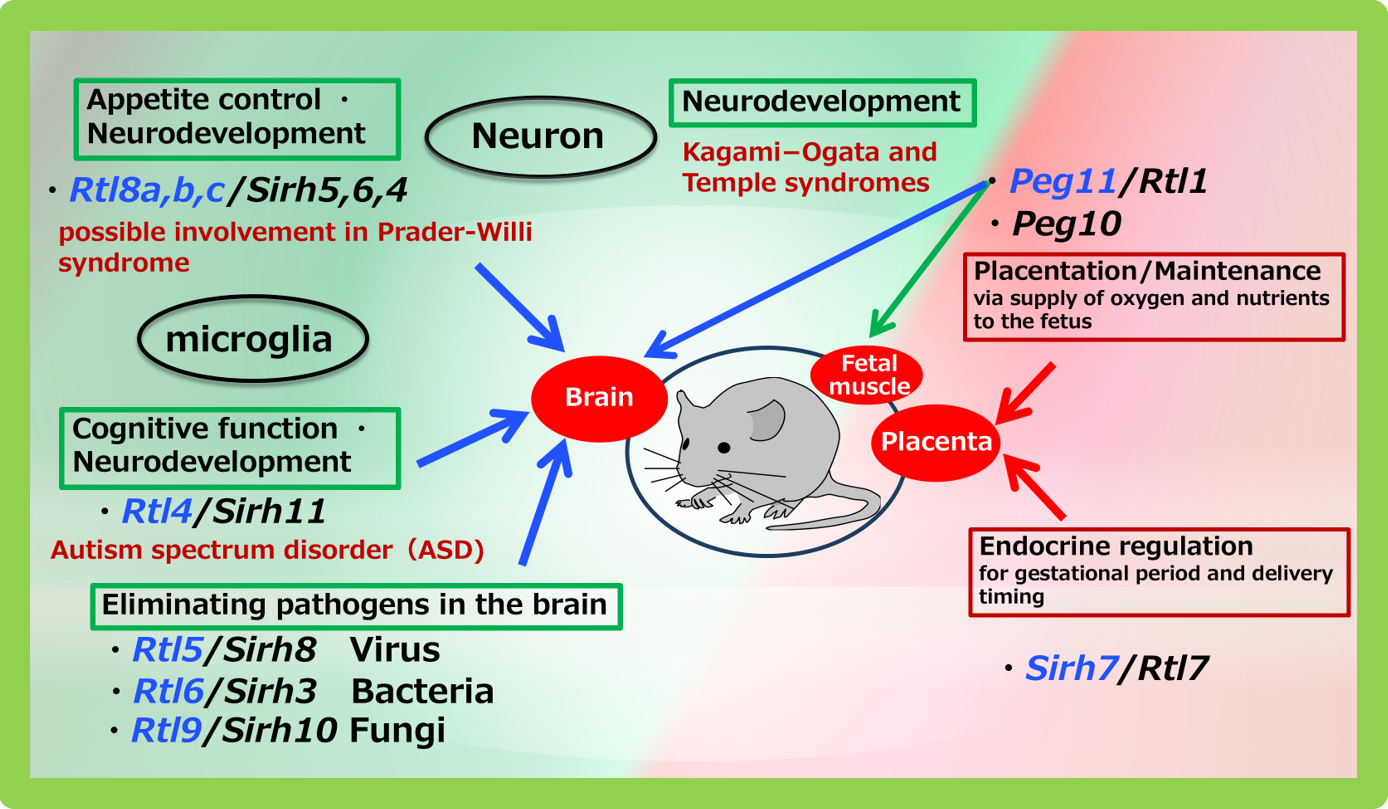
Figure 3 summarizes the function of the 10 of the 11 SIRH/RTL genes whose roles have been elucidated. The overall picture shows that virus-derived acquired genes play key roles in the placenta and brain, two organs that are characteristic of mammals.
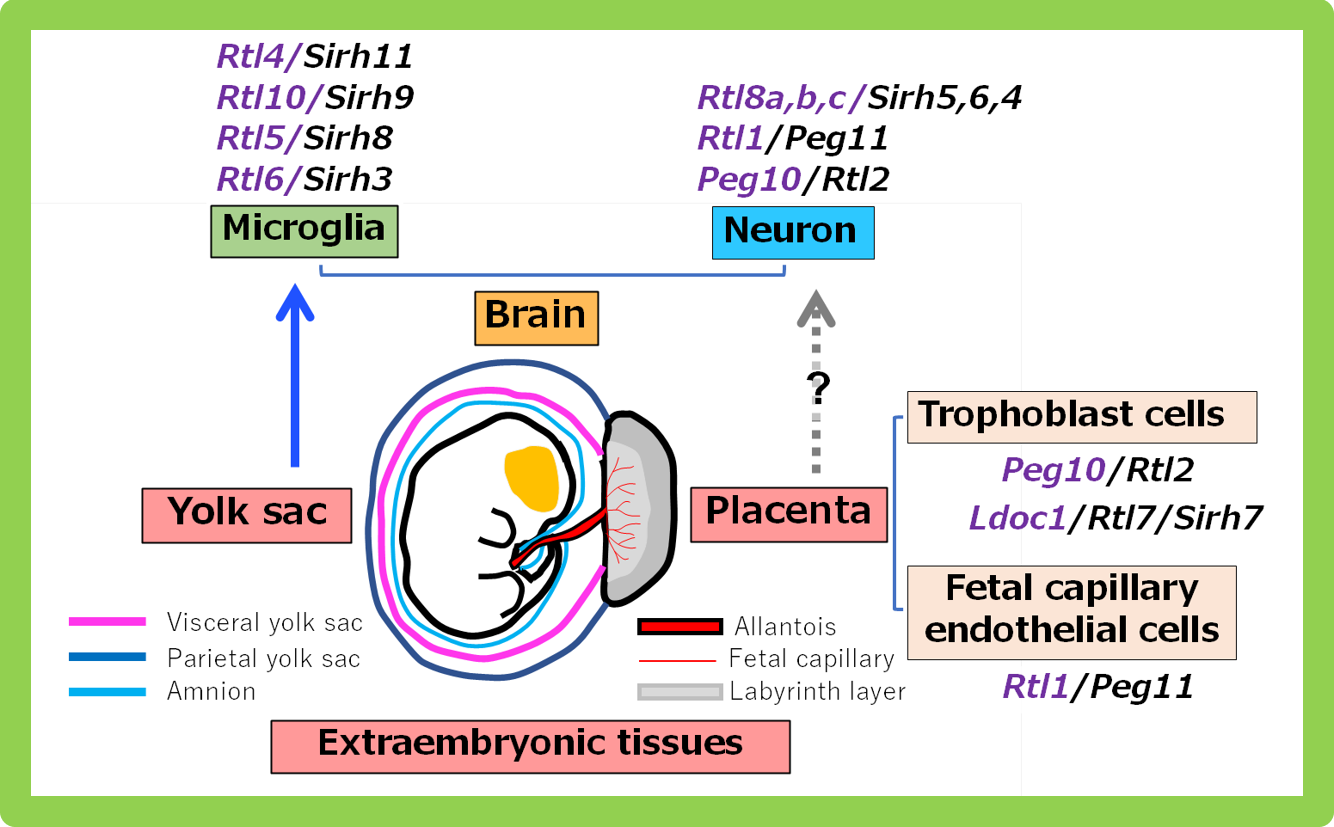
Although the placenta and brain are distinct organs, they share an intriguing connection through extraembryonic tissues (non-fetal tissues). Unlike other brain cells, microglia originate from the yolk sac early in development before migrating to the fetal brain. As a result, the acquired SIRH/RTL genes are active either in the placenta or in yolk sac-derived microglia.
We believe this underscores the importance of the extraembryonic tissues of the placenta and yolk sac, where these genes are acquired. We propose the following new hypothesis: The placenta, yolk sac and other extraembryonic tissues serve as sites of gene acquisition. These tissues, which have lower DNA methylation levels than the fetus—where viral genes are completely repressed by high DNA methylation—do not fully suppress viral genes that have integrated into the genome. This ‘leaky’ expression allows for the selection and acquisition of new genes when mutations confer beneficial functions to the host, particularly through functions in extraembryonic tissues. In essence, extraembryonic tissues act as sites for evolutionary testing, facilitating the evolution of eutherians (Fig. 4).
Thus, genes that function in the placenta or originate in the yolk sac were acquired first. Subsequently, placental genes, such as Peg10 and Peg11/Rtl1, may have also acquired functions in neurons, although how these genes came to play a role in neuronal function remains unresolved. Ultimately, the genes acquired in extraembryonic tissues may have accelerated eutherian brain evolution from two directions: one through neurons and the other through microglia. As discussed in more detail in Theme 10, at least five virus-derived genes are involved in placentation in eutherians. It is intriguing that the newly formed placenta may have served as a site for the acquisition of additional new genes, further accelerating the evolution of eutherians.
The SIRH/RTL genes most likely became new genes in mammals as a result of foreign (virus-derived) DNA inserting the genome in our mammalian ancestors, having novel functions through multiple mutation, and natural selection for its beneficial properties during evolution. The present “necessity” in life is partly supported by these acquired genes of virus origin that were inserted into the genome by “chance” and, through natural selection, became genes with beneficial functions. We believe that our research provides a concrete example of how acquired genes of viral origin have contributed to the large-scale evolution of mammals.
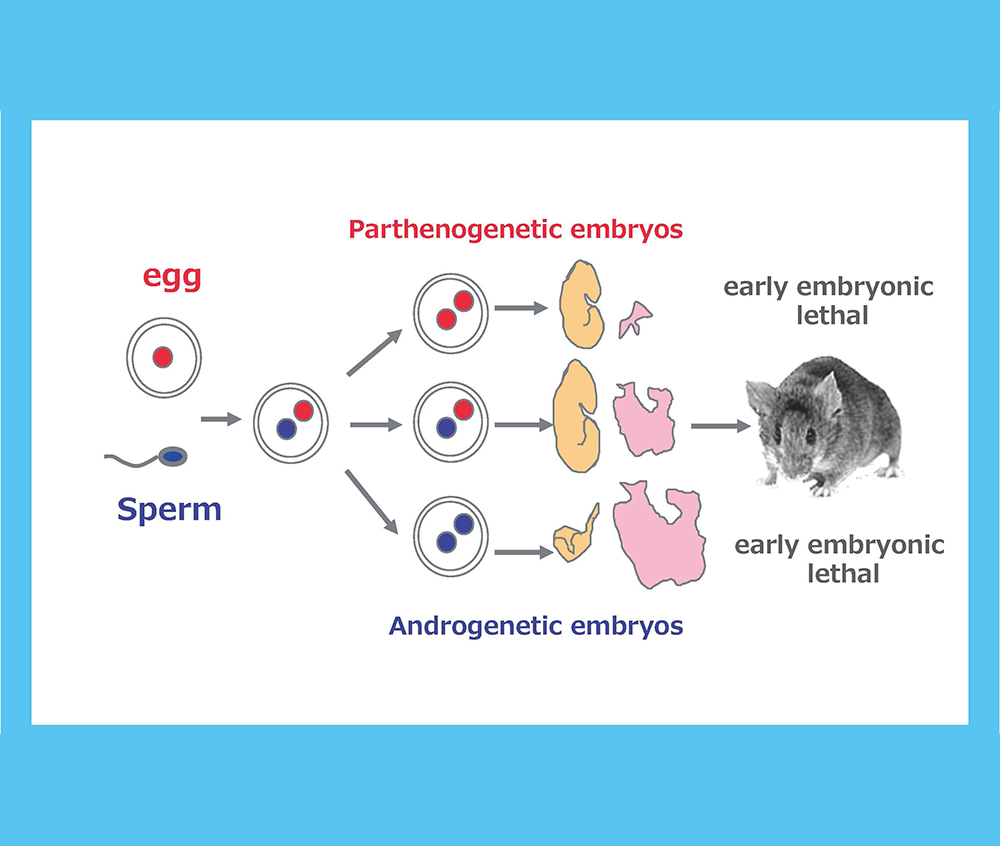
Genomic imprinting is a mammalian-specific epigenetic mechanism regulating the parent-of-origin-specific expression of a subset of genes. It was independently discovered in 1984...
We returned to Japan with materials provided by Azim Surani, such as parthenogenetic embryos and control, normally fertilized embryos...
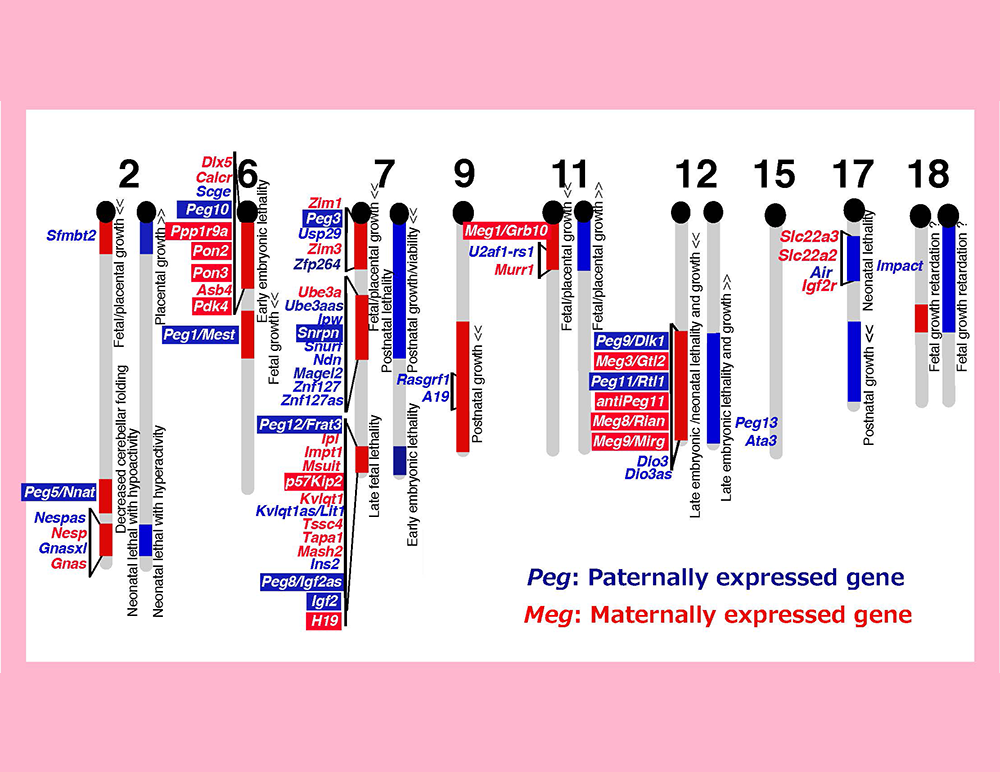
The imprinted regions are comprised of clusters of imprinted genes, meaning that the imprinted genes are regulated at the regional level, not in a gene-by-gene manner. Each imprinted region...
We assumed that the imprinted gene essential for placenta formation would exist in the imprinted region on mouse proximal chromosome 6 because this is the only imprinted region...
Maternal duplication of the distal chromosome 12 causes late stage embryonic/ neonatal lethality...
Why has genomic imprinting been so widely conserved in mammals? Genomic imprinting is seemingly contradictory...
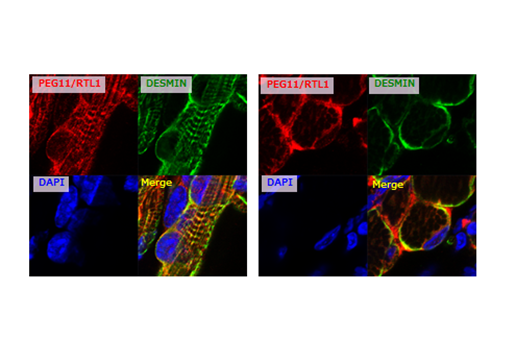
In Theme 5, we showed that Peg11/Rtl1 plays an essential role in the maintenance of placental...
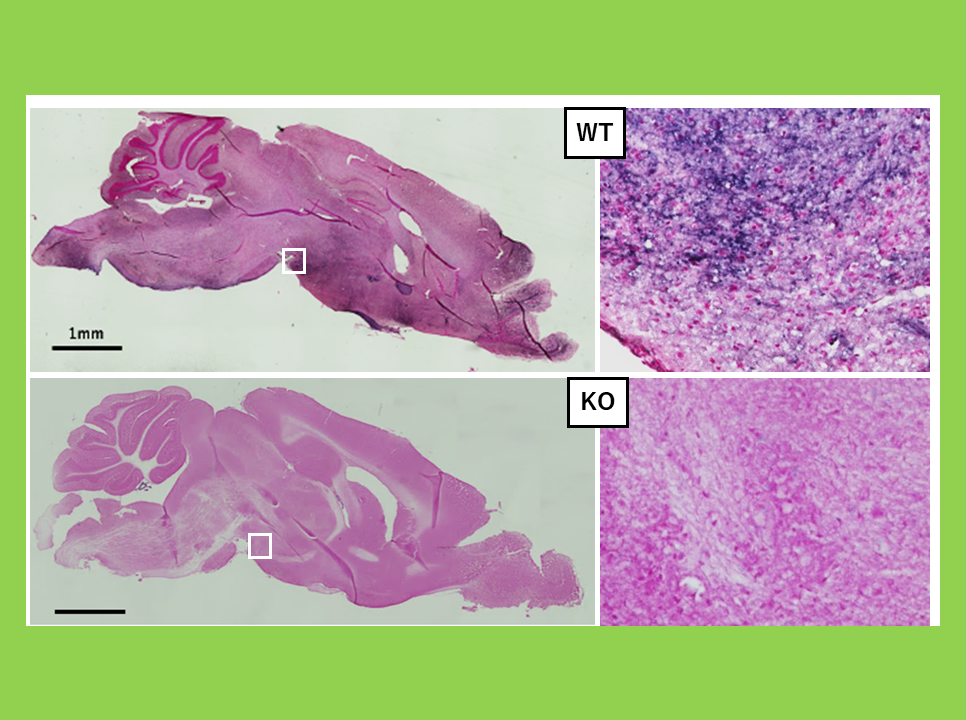
Among the SIRH/RTL genes functioning in neurons, PEG11/RTL1is known as the causative gene for Kagami-Ogata syndrome and Temple syndrome...
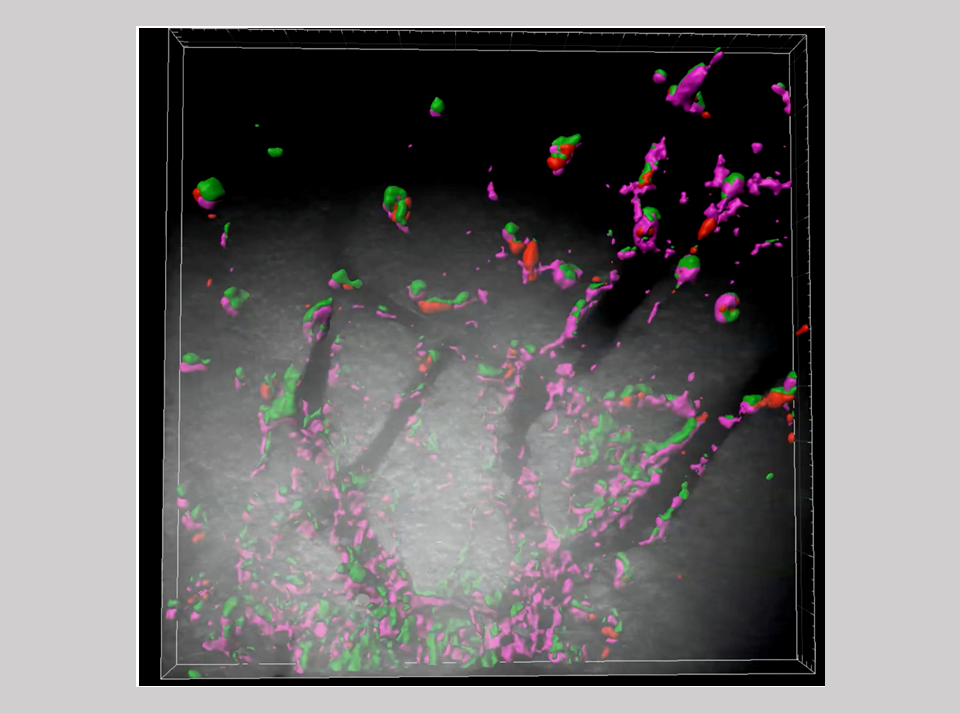
Innate immunity is a fundamental biological defense system, widely conserved across the animal kingdom...
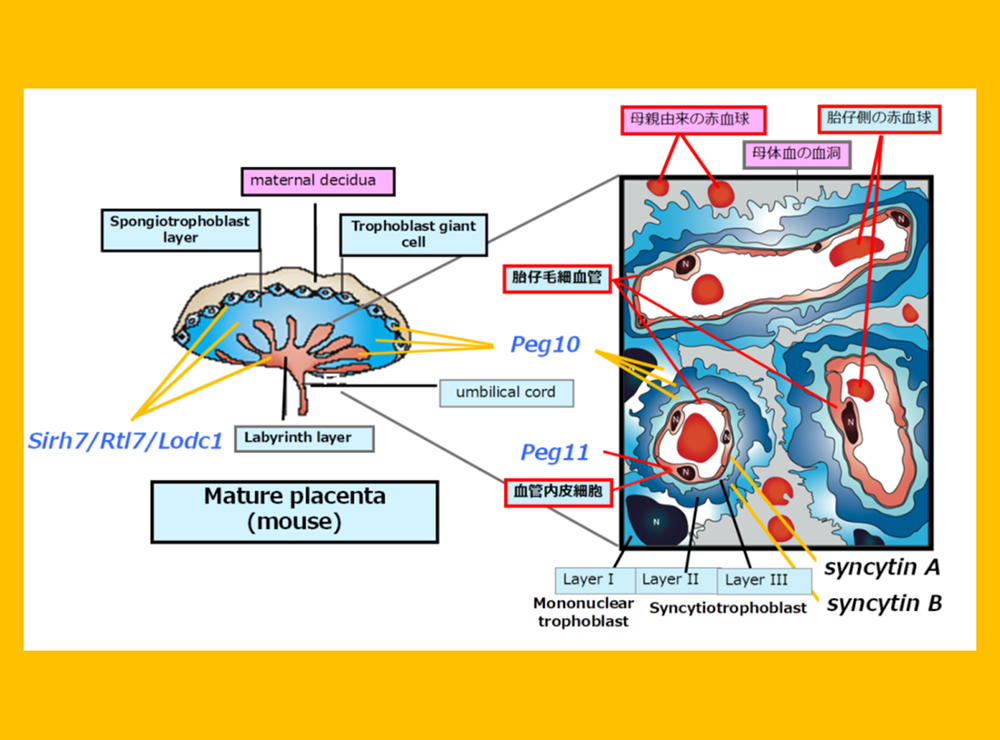
“The placenta”serves as a fascinating example of an innovative evolution, where virus-derived genes have been...